Description of the Trichoptera data set
PLN team
2025-11-03
Source:vignettes/Trichoptera.Rmd
Trichoptera.RmdPreliminaries
This vignette describes the data set trichoptera, which
is used in all examples, tests and other vignettes of the package
PLNmodels. This data set regroups the main striking
characteristics of ecological count data (encompassing tables of
abundances and covariates). The low dimensional space of the data also
ensures that it is well suited for illustrative purposes.
The packages required to run the vignette are the following:
The trichoptera data set
The order Trichoptera (or caddisflies) are a group of insects with aquatic larvae and terrestrial adults. The ecological data set trichoptera (Usseglio-Polatera and Auda 1987) describes abundances of Trichoptera species (hereafter the counts), accompanied with some meteorological factors (hereafter the covariates) that may influence their presence during the sampling1.

Macronema Zebratum captured by Y. Dubuc at Donacona (Québec), 06-20-2001.
The data is directly available once PLNmodels is
loaded. Comprehensive information and description are available to the
user with ?PLNmodels::trichoptera.
data(trichoptera)Formatting
Data are originally stored in a list of two data frames for abundances and covariates. We first prepare the data see the corresponding vignette for easy use in the multivariate framework of PLNmodels. Offsets are automatically computed:
trichoptera <- prepare_data(trichoptera$Abundance, trichoptera$Covariate)Data are now stored in the trichoptera data frame which
includes 49 rows (the observations - or trapping nights) and 9 columns.
As can be seen, there are 2 multivariate columns (matrices of counts and
offsets) and 7 univariate columns (vectors of covariates):
str(trichoptera)## 'data.frame': 49 obs. of 9 variables:
## $ Abundance : num [1:49, 1:17] 0 0 0 0 0 0 0 0 0 0 ...
## ..- attr(*, "dimnames")=List of 2
## .. ..$ : chr [1:49] "1" "2" "3" "4" ...
## .. ..$ : chr [1:17] "Che" "Hyc" "Hym" "Hys" ...
## $ Temperature : num 18.7 19.8 22 23 22.5 23.9 15 17.2 15.4 14.1 ...
## $ Wind : num -2.3 -2.7 -0.7 2.3 2.3 -2 -4.7 -1 -2.7 -3.7 ...
## $ Pressure : num 998 1000 997 991 990 ...
## $ Humidity : num 60 63 73 71 62 64 93 84 88 75 ...
## $ Cloudiness : num 19 0 6 81 50 50 100 19 69 6 ...
## $ Precipitation: num 0 0 0 0 0 0 1.6 0 1.6 0 ...
## $ Group : Factor w/ 12 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ Offset : num 29 13 38 192 79 18 8 34 12 4 ...We thus rely on a not so common use of the data.frame
structure, where a column may be a matrix and not necessarily an atomic
vector. This formatting is very handy for model specification with
R formulas, especially in a multivariate
setting like in PLNmodels.
Table of counts (abundancies)
The Abundance column is a
matrix of abundancies (or counts) for the 17 species found during the 49
trapping nights.
| Che | Hyc | Hym | Hys | Psy | Aga | Glo | Ath | Cea | Ced | Set | All | Han | Hfo | Hsp | Hve | Sta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 5 | 0 | 17 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 2 | 0 | 1 |
| 0 | 0 | 3 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| 0 | 0 | 1 | 0 | 32 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 1 | 0 | 0 |
| 0 | 0 | 3 | 0 | 176 | 4 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 3 | 0 | 2 |
| 0 | 0 | 4 | 0 | 69 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 2 |
| 0 | 0 | 2 | 0 | 14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
In all other vignettes and journal papers associated with
PLNmodels, the count table is denoted by
in the mathematical model and Y in the R
environment.
A basic representation of the matrix of counts (here transposed and log-transformed), shows the typical huge dispersion between low and highly abundant species.
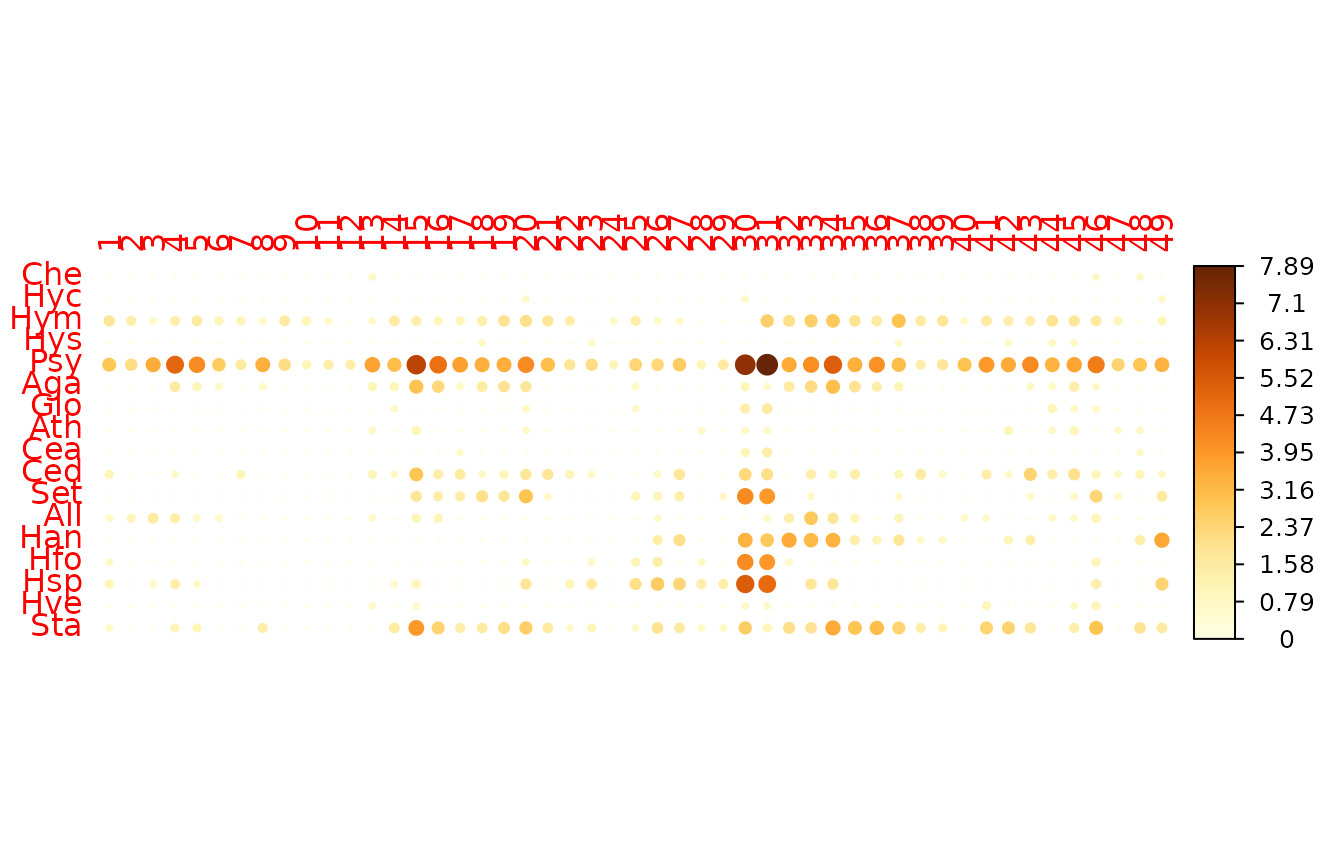
log-counts in the trichoptera data set
Covariates (external meteorological effect, groups)
Additional information was collected during the sampling, which corresponds to external covariates the effect of which may or may not be taken into account in the model (depending on the question at play). In the trichoptera data set, those covariates correspond to meteorological factors plus a categorical variable indicating the family of the caughts specimens.
| Temperature | Wind | Pressure | Humidity | Cloudiness | Precipitation | Group |
|---|---|---|---|---|---|---|
| 18.7 | -2.3 | 998.5 | 60 | 19 | 0 | 1 |
| 19.8 | -2.7 | 999.5 | 63 | 0 | 0 | 1 |
| 22.0 | -0.7 | 997.2 | 73 | 6 | 0 | 1 |
| 23.0 | 2.3 | 991.1 | 71 | 81 | 0 | 1 |
| 22.5 | 2.3 | 990.1 | 62 | 50 | 0 | 1 |
| 23.9 | -2.0 | 990.1 | 64 | 50 | 0 | 1 |
The design matrix arising from the covariates is denoted by
in our mathematical model and X within the R
environment.
Offsets and the compositionality issue
A common issue with (microbiological) ecological data is the compositionality problem: counts can only be compared to each other within a sample but not across samples as they depend on a sample-specific size-factor, which may induce spurious negative correlations of its own. Besides, the sampling of some particular species may be biased, for instance when different technologies are used to sample different types of species. Those technical biases can be encoded in a table of offsets. In the case at hand, we have a natural offset for each sample that corresponds to the total counts per night, specified by an numeric of offsets. Note that the offset term remains the same in a given sample albeit sometimes one might include an offset specific to both the sample and the species. The formula syntax accepts either no offset, a vector or a matrix or specify the offsets term.
Here, we have a vector whose corresponding column is named
Offset in in the trichoptera data frame:
trichoptera$Offset## [1] 29 13 38 192 79 18 8 34 12 4 4 3 49 33 600
## [16] 172 58 51 56 127 35 13 17 3 27 40 44 8 9 1599
## [31] 2980 88 135 327 66 90 63 15 14 20 70 53 95 43 62
## [46] 149 16 31 86See the importation vignette and the
function prepare_data() to learn more about how Offset can
be computed in PLNmodels.
Offsets are denoted by
in the mathematical model across other vignettes, and by O
in the R environment.
References
The original data set is available in the ade4 package. We consider here a different version where we only keep a subset of the original meteorological features, for illustrative purposes↩︎